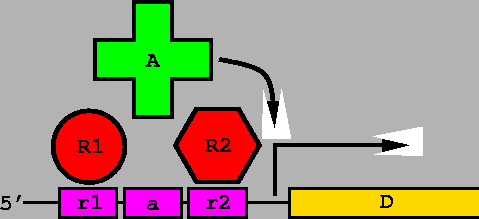
 An information-theory based model of Fis protein DNA binding was searched across the E. coli genome. The distances between the zero coordinates of successive sites were recorded and tabulated in this graph. Red curve: search of E. coli genome; green curve: search over equiprobable random sequence; blue curve: mathematical model for randomly placed sites. This model was constructed by considering a genome of size G=4639221 bases (the U00096 E. coli K-12 MG1655 genome) having n=154112 sites (Ri > 0 bits) so that the probability of a site being at one position is p = n/G. Then the number of sites with separation d is Gp2(1-p)d. Similar results are obtained for a 2.5 bit cutoff, the lowest observed Fis site in our set. Arrows indicate spacings of 7 and 11 base pairs. |
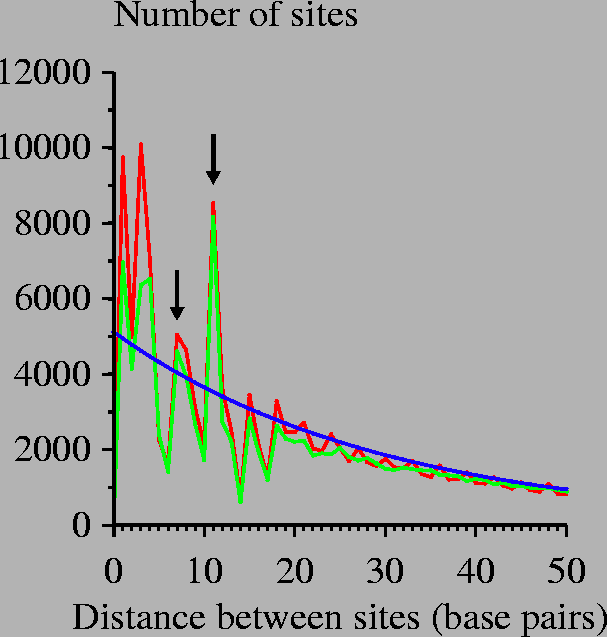
 The sequence logo for Fis [19,5] is shown three times. The upper and lower logos are shifted +11 and +7bases to the right (respectively) relative to the middle logo. Dashed waves indicate the phase of the shifted site; solid waves indicate the phase of the unshifted site. The in-phase sine waves, with a wavelength of 10.6 bases, show that Fis sites shifted by 11 bases would be on the same face of the DNA [28,101,102], while the out-of-phase waves of Fis sites shifted by 7 bases indicate binding to opposite faces. Arrows are at positions where the logo is self-similar after a shift. Red arrows (pointing downwards from the +11 shift) mean that the contacts by Fis to the bases would interfere because they would be on the same face of the DNA. Green arrows (pointing upwards from the +7 shift) mean that the contacts could be simultaneous because they are on opposite faces. In a sequence logo, the height of each letter is proportional to the frequency of the corresponding base at that position in the sites, and the height of the stack of letters represents the sequence conservation in bits. For clarity, the sine waves run from 1 to 1.6 bits. |
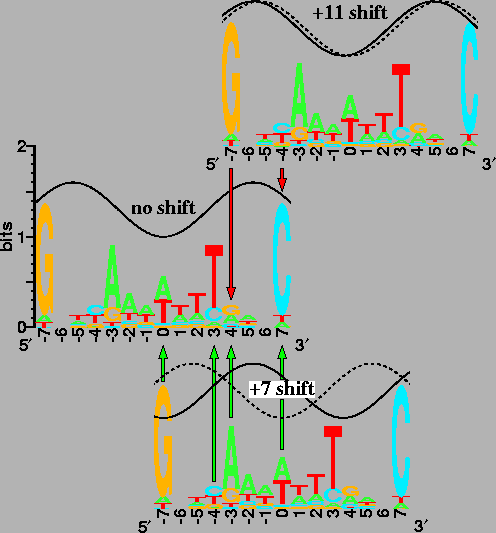
 (a) A single Fis dimer binding to DNA. (b) Two Fis dimers binding to Fis sites separated by 11 base pairs. (c) Two Fis dimers binding to Fis sites separated by 7 base pairs. The DNA backbone is color coded: A: green, C: blue, G: orange, T: red. The models of Fis interacting with DNA were built using Insight II software from Biosym Technologies, Inc., on an IRIS computer (Silicon Graphics, Inc.), and displayed with RasMol 2.5, available at http://molbiol.soton.ac.uk/rasmol.html or ftp://ftp.dcs.ed.ac.uk/pub/rasmol/. The Fis protein coordinates are those of the Protein Data Bank (http://www.rcsb.org/pdb/) entry 1fia. (See Materials and Methods for further details.) |
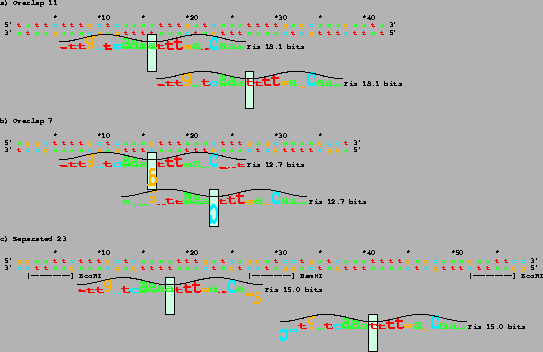 The predicted Fis sites are shown by sequence walkers
floating below each self-complementary DNA sequence
[10,5].
In a walker, the vertical
green box marks the zero base of the binding site.
The box also shows the vertical scale, with the upper edge being at +2 bits
and the lower edge being at -3 bits.
The height of each letter
is determined from the bit value
in the
individual information weight matrix
[21,10,5].
Negative weights are represented by drawing the letter
upside-down
and placing it below the zero bit level.
To indicate predicted relative orientations,
the peaks of sine waves correspond to where Fis
would bind into the major groove.
Three DNAs were designed, each having
two Fis sites spaced 11, 7 and 23 bases apart.
Design details are given in Materials and Methods.
The total strength of a site is the sum of the information weights for each
base.
The 18.1 bit Fis sites are
3.4
standard deviations
higher than the average Fis site
in natural sequences [5,21].
The
12.7 and 15.0 bit sites are
1.6 and 2.4
standard deviations above average (respectively).
The predicted Fis sites are shown by sequence walkers
floating below each self-complementary DNA sequence
[10,5].
In a walker, the vertical
green box marks the zero base of the binding site.
The box also shows the vertical scale, with the upper edge being at +2 bits
and the lower edge being at -3 bits.
The height of each letter
is determined from the bit value
in the
individual information weight matrix
[21,10,5].
Negative weights are represented by drawing the letter
upside-down
and placing it below the zero bit level.
To indicate predicted relative orientations,
the peaks of sine waves correspond to where Fis
would bind into the major groove.
Three DNAs were designed, each having
two Fis sites spaced 11, 7 and 23 bases apart.
Design details are given in Materials and Methods.
The total strength of a site is the sum of the information weights for each
base.
The 18.1 bit Fis sites are
3.4
standard deviations
higher than the average Fis site
in natural sequences [5,21].
The
12.7 and 15.0 bit sites are
1.6 and 2.4
standard deviations above average (respectively). |
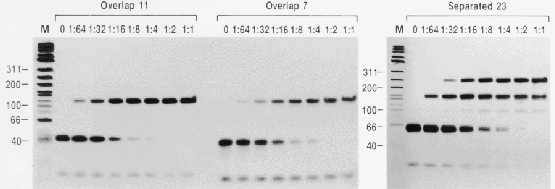 Each lane contains increasing
concentrations of Fis protein,
beginning with
no Fis, Fis diluted 1 to 64, etc.
The 1:1 dilution was at 2200 nM Fis.
This concentration was chosen intentionally so that with
the 1 nM of DNA used in this experiment,
the protein/DNA ratio was 2-fold higher
than that
needed to strongly shift
DNA containing
the 8.9 bit wild-type hin distal Fis site
[29].
The sequences are given in Fig. 4.
Marker lanes (M) contain 10 ng of biotinylated Each lane contains increasing
concentrations of Fis protein,
beginning with
no Fis, Fis diluted 1 to 64, etc.
The 1:1 dilution was at 2200 nM Fis.
This concentration was chosen intentionally so that with
the 1 nM of DNA used in this experiment,
the protein/DNA ratio was 2-fold higher
than that
needed to strongly shift
DNA containing
the 8.9 bit wild-type hin distal Fis site
[29].
The sequences are given in Fig. 4.
Marker lanes (M) contain 10 ng of biotinylated |
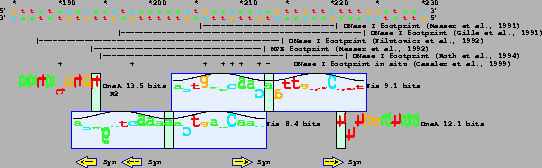 Sequence data are from GenBank accession K01789
[103].
The horizontal dashes below the sequence
represent regions protected by Fis.
Locations of DnaA sites are from [104]
and Fis footprint data are from
[104,52,66,54,53,105].
The asymmetric
DnaA individual information matrix was created from 27 experimentally
demonstrated DnaA binding sites
[102].
DNA synthesis start sites are indicated by yellow arrows and `Syn'
[64],
however start sites
have also been mapped to the left side of oriC
[69].
Blue boxes mark two Fis sites separated by 11 bases.
DnaA site directionality is indicated by letters turned
sideways in the direction that
DnaA binds [10].
Sequence data are from GenBank accession K01789
[103].
The horizontal dashes below the sequence
represent regions protected by Fis.
Locations of DnaA sites are from [104]
and Fis footprint data are from
[104,52,66,54,53,105].
The asymmetric
DnaA individual information matrix was created from 27 experimentally
demonstrated DnaA binding sites
[102].
DNA synthesis start sites are indicated by yellow arrows and `Syn'
[64],
however start sites
have also been mapped to the left side of oriC
[69].
Blue boxes mark two Fis sites separated by 11 bases.
DnaA site directionality is indicated by letters turned
sideways in the direction that
DnaA binds [10]. |
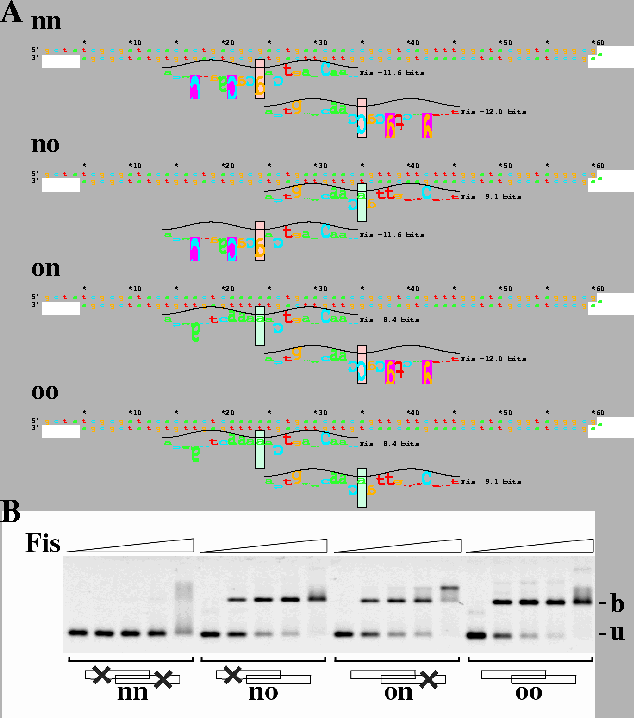 A. Design of wild-type and mutated Fis sites from
E. coli oriC.
Four hairpin oligos were designed and designated
nn,
no,
on
and
oo
where n means no site
because of engineered mutations
(pink boxes, with information less than zero)
and o means that there is
a complete wild-type origin Fis site
(green boxes, with positive information).
For example,
no contains only the Fis site closest to R3 on the right side.
B. Gel mobility shift assay with oriC sites using the oligos
shown in part A at a concentration of 10 nM each.
Fis concentrations were 0, 30, 100, 300 and 1000 nM.
u: unbound DNA;
b: Fis bound DNA.
A. Design of wild-type and mutated Fis sites from
E. coli oriC.
Four hairpin oligos were designed and designated
nn,
no,
on
and
oo
where n means no site
because of engineered mutations
(pink boxes, with information less than zero)
and o means that there is
a complete wild-type origin Fis site
(green boxes, with positive information).
For example,
no contains only the Fis site closest to R3 on the right side.
B. Gel mobility shift assay with oriC sites using the oligos
shown in part A at a concentration of 10 nM each.
Fis concentrations were 0, 30, 100, 300 and 1000 nM.
u: unbound DNA;
b: Fis bound DNA. |
|
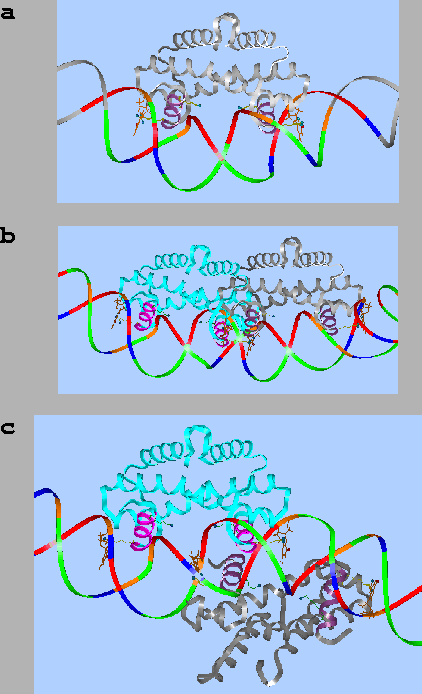
An activator protein molecule A (green plus) binds to a DNA molecule at position a. When the activator binds, it turns on the promoter for gene D. Two repressor protein molecules R1 and R2 (red circle and red hexagon respectively) bind to DNA at positions r1 and r2. Binding to either r1 or r2 interferes with binding by A, so the activator can only bind when the two repressors are absent. Assigning the presence of a molecule as `1' or `true' and the absence as `0' or `false', then D = R1 NOR R2. By connecting such NOR gates together, any computer circuit can be built. |
![]()