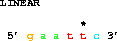 To get the first 6 bases (containing just the EcoRI site)
we say:
To get the first 6 bases (containing just the EcoRI site)
we say:
Terminiology used is described in a glossary.
The concept of the Delila system is to extract fragments of sequence from a library (database) of sequences before beginning any analysis of the sequences. This has a number of advantages, including automating the analysis process, avoiding editing sequences (which will lead to mistakes!), the ability to permanently record the sequences used in a compact form (instructions) and therefore the ability to repeat an analysis. The extraction is done by a librarian program named Delila. One gives Delila instructions for what fragments to obtain and how to mutate them. The returned result given by the librarian is -- of course! -- a book.
An important feature of Delila is that the coordinate system of each sequence in the book corresponds to that in the parent library. This way you won't go crazy trying to figure out the locations of bases - all output has the same coordinate system. (The exception is if you make mutations, in which case coordinates get renumbered on the 'downstream' side.)
If you already have a Delila Library (i.e. the 6 files lib1, lib2, lib3, cat1, cat2, cat3) then you can skip this section. If not, you need to create one.
The first step is to create a Delila book containing the genomic or artificial sequence you want to manipulate. There are a number of programs you can use to do this:
Next, you need to create the Delila library. In a Unix system:
cp book l1 # copy your book to the file l1
touch l2 l3 catalp # make empty files
catal # run the catal program
Delila will now run using the 6 library files and an instruction file.
Since Delila produces a book, it is natural that the first instruction in a set of Delila instructions is the title to be given to the book:
title "An example book";
Note that delila will accept both single (') and double (") quotes.
You can have any title you like. I would, however, recommend this format:
title 'Fis sites version = 1.81 of fis.inst 2002 Apr 24';This includes four important components:
Next, the desired source sequence must be specified. Delila was built before GenBank existed and it assumes that the database is organized by organism and chromosome (as opposed to the current mess of entries). So one defines these:
organism H.sapiens;
chromosome H.sapiens;
Next one needs to choose the particular sequence of DNA, called a piece:
piece LINEAR;where LINEAR would usually be the GenBank ACCESSION number.
Having specified the sequence we want, we now can make a series of requests to get particular parts of the sequence. Suppose that the wild-type sequence named LINEAR begins with the EcoRI site 5' gaattc 3', with bases numbered 1 to 180. Then to obtain the entire sequence we can say:
get all piece;
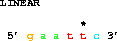 To get the first 6 bases (containing just the EcoRI site)
we say:
To get the first 6 bases (containing just the EcoRI site)
we say:
get from 1 to 6;The lister program puts an asterisk ('*') every 5th base, and numbers every 10th base. (This way you won't go crazy counting bases - you never need to count more than 3 positions to identify a base.)
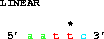 To get the second to sixth bases one can say:
To get the second to sixth bases one can say:
get from 2 to 6;which gives 5' aattc 3'.
 One can also get the complement:
One can also get the complement:
get from 6 to 2 direction -;which also gives 5' gaatt 3'. Note that the asterisk in the figure is still over base 5. Delila retains the original coordinate system, which means that you can compare output from different extractions and the coordinates of the bases remain the same.
Here's a puzzler:

get from 2 to 5 direction +;
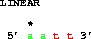
get from 5 to 2 direction -;Why are these the same?
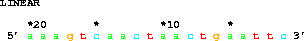 An example longer sequence is:
An example longer sequence is:
get from 20 to 1 direction -;giving 5' aaagtcaactaactgaattc 3', which shows how the coordinate system decreases. (Note the EcoRI site at the 3' end.)
Having obtained the sequence(s) we want, Delila's job is over. Other programs are used to display and analyze the sequence. For these examples I used the Lister program for the figures. Lister gives the sequence, carefully labeled with 5' and 3' on the ends. Every 5th base is marked by an asterisk, and every 10th base is numbered. This way you will never need to count more than 3 bases to determine the coordinate of any base.
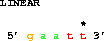 A powerful way to get sequences is relative to a particular point:
A powerful way to get sequences is relative to a particular point:
get from 3 -2 to 3 +2;which gets 2 bases before coordinate 3 to 2 bases after coordinate 3, that is from base 1 to base 5: 5' gaatt 3'. Generally one does not want to repeat the second coordinate, so one can use the
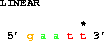
get from 3 -2 to same +2;where 'same' refers to the coordinate given after the word 'from'. This is the most convenient form for specifying binding site locations. For more examples, see: Making Delila Instructions for Symmetric Sites.
There are three ways to make changes.
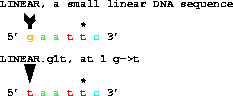 1. A CHANGE requires the previous base, the coordinate to
change and then the new base:
1. A CHANGE requires the previous base, the coordinate to
change and then the new base:
get from 1 to 6 with g1t;
gives taattc. The base that changes from a G at 1 to an T is marked by the tail and head of an arrow. The figure is produced by first running Delila to extract the sequence(s) and to produce the marking information. This information is then used by Lister to create the postscript.
How do I write my instructions if I want
the complementary sequence?
Glad you asked.
Coordinates of changes are always given on the original wild-type coordinate
system. The rule is:
The coordinates given in the mutation and the sequences given refer always to the sequence written 5' to 3' in the *positive* coordinate direction.The reason for doing things this way is that you would go absolutely crazy if you had to change the definition of the mutation merely if you wanted the complementary sequence!
 For example, starting again from 5' gaattc 3':
For example, starting again from 5' gaattc 3':
get from 6 to 1 with g1t;Delila makes the mutation and then complements the sequence to give 5' gaatta 3'. Note that the first sequence in the illustration is already complemented. You can see this because the asterisk ('*') marks the 5th base.
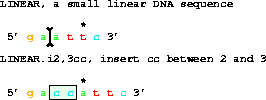 2. An INSERTION uses two coordinates and a sequence. The sequence
BETWEEN the coordinates is removed and the given sequence is inserted.
2. An INSERTION uses two coordinates and a sequence. The sequence
BETWEEN the coordinates is removed and the given sequence is inserted.
get from 1 to 6 with i2,3cc;gives gaccattc.
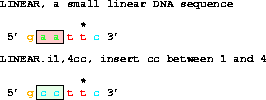 Changing that to:
Changing that to:
get from 1 to 6 with i1,4cc;does a replacement to give gccttc.
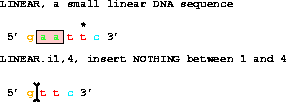 Finally,
Finally,
get from 1 to 6 with i1,4;deletes to give gttc.
Note that any change can be made with this definition; the other methods are available for convenience.
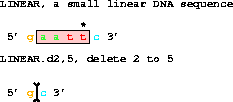 3. A DELETION takes two coordinates. The sequence INCLUDING
the coordinates is removed.
3. A DELETION takes two coordinates. The sequence INCLUDING
the coordinates is removed.
get from 1 to 6 with d2,5;gives gc. Coordinates outside the end of the piece are allowed.
 Combined changes are possible. Separate the
changes with periods:
Combined changes are possible. Separate the
changes with periods:
get from 1 to 6 with g1t.i1,4cc;gives tccttc.
title "ABCR mutation";
organism H.sapiens;
chromosome H.sapiens;
set doubling on;
piece Y15651;
name "mutation at exon 17 acceptor";
get from 63 -25 to same +7 with g64a;
Two new commands are introduced here:
set doubling on;which tells Delila to give both the original sequence and the sequence with the mutation and
name "mutation at exon 17 acceptor";which tells Delila to name the new sequence. The result, when displayed by the lister program, is:
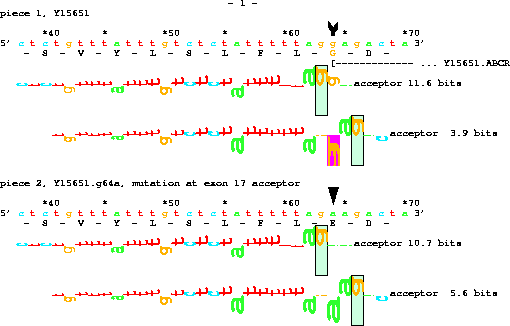
In all of the examples above, the book was given to the lister program, which generated PostScript output. Lister has a special mode for displaying sequences along with their mutations: the 'pagetrigger' parameter is set to 'd'. To use this feature, create a mutation instruction using 'with' and be sure to 'set doubling on' before that point in the instructions. In the book Delila will put the original sequence along with the mutation sequence. Delila will also create a 'marksdelila' file which contains information about how to mark the mutation. Append the 'marksdelila' file to the end of the arrow definition file (marks.arrow) and run lister:
The resulting 'map' file is in PostScript and can be sent to a printer, displayed on your screen or converted to PDF.delila cat marks.arrow marksdelila > marks lister
The Delila language provides two ways to create comments in the instruction files. Both are 'Pascal-like' since the same form is used in the computer language Pascal:
(* Two character comments *)and
{ One character comments }Material inside comments is ignored by Delila. Comments of one type can be nested inside the other type. I commonly make my comments using (* and *) and then use the braces { and } to block off instructions I don't want temporarily.
I strongly recommend putting in the date and the file name in the title, and at least a short description of what the instruction set is about in a comment. It is also useful to add citations for evidence that the sequence is a binding site, and to mention the kind of data that supports this (e.g. footprinting, gel shift assay, mutations).
Binding sites can have three kinds of symmetry, as discussed in the glossary entry on binding site symmetry. The corresponding Delila instructions are of increasing difficulty:
asymmetric. There is one instruction per site:
get from 2718 -20 to same +30 direction +; get from 3141 -20 to same +30 direction +; get from 6931 +20 to same -30 direction -;Note how the last one would be in the opposite orientation relative to the first two.
odd symmetric. There are two instructions per site. The second instruction switches the direction but keeps the 'from' coordinate the same:
get from 2718 -20 to same +20 direction +; get from 2718 +20 to same -20 direction -; get from 3141 -20 to same +20 direction +; get from 3141 +20 to same -20 direction -; get from 6931 -20 to same +20 direction +; get from 6931 +20 to same -20 direction -;Note how the pattern of the ranges switches between positive and negative.
even symmetric. There are two instructions per site. The second instruction switches the direction AND CHANGES the 'from' coordinate by one base. This follows our convention that the center of symmetry is between bases 0 and 1:
get from 2718 -20 to same +20 direction +; get from 2719 +20 to same -20 direction -; get from 3141 -20 to same +20 direction +; get from 3142 +20 to same -20 direction -; get from 6931 -20 to same +20 direction +; get from 6932 +20 to same -20 direction -;Note 1: how the pattern of the ranges switches between positive and negative.
Note: the ranges given above are only examples. We generally take a very large range such as -200 to +200 for our initial analysis to get a feeling for the background noise of the information curve.
Delila has a number of parameters that have preset values which you can change. You can use the word 'default' or 'set' to change them.
set numbering piece; (* number the pieces *) set numbering 1; (* start numbering at 1 *) set numbering off; (* turn off or on numbering *) set numbering all; (* number all book parts *)
set out-of-range reduce-range; (* reduce to the nearest end *) set out-of-range halt; (* stop *) set out-of-range continue; (* keep on going *)
set coordinate zero; (* the 'from' base becomes coordinate zero
in the resulting book *)
For example, the instruction 'get from 20 -10 to same +10;'
results in a new coordinate system that runs from -10 to +10.
set coordinate 5; (* the 'from' base becomes coordinate 5
in the resulting book *)
set coordinate normal; (* return to using the original coordinates *)
set doubling on; (* double pieces *)An example is Medical Applications of Sequence Walkers: ABCR Mutation G863A.
set arrowlength 1.5; (* Default arrow length is just a triangle *)Examples are shown above.
If you would like to know more about the Delila language, then you can look at the LIBrary DEFinition, LIBDEF.
The delila system has a number of ways to automatically generate delila instructions:
![]()

Schneider Lab
origin: 1999 May 2
updated:
version = 2.05 of delilainstructions.html 2009 Jan 27
![]()