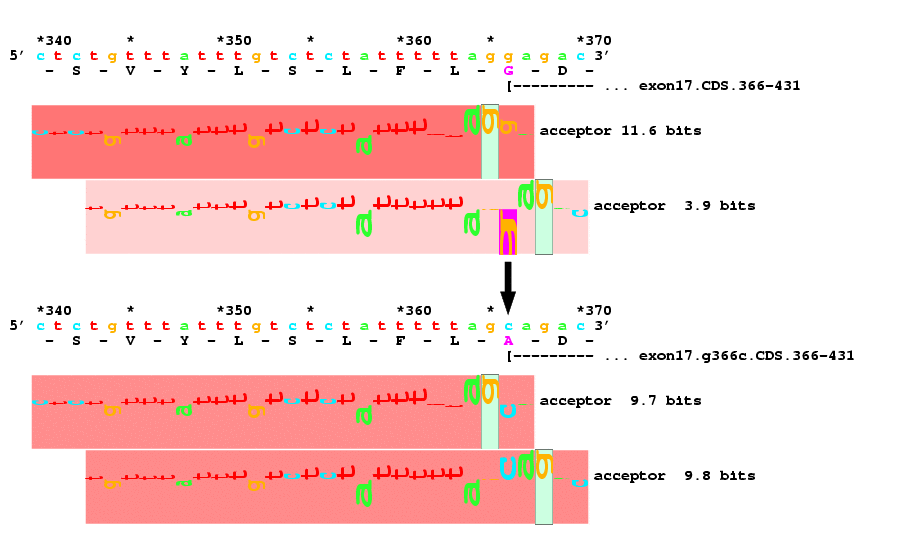

The ABCR protein is a putative retinoid transporter in rod photoreceptor cells, alterations in which cause numerous inherited eye diseases. A G to C mutation at position 366 (arrow) of the ABCR gene is responsible for Stargardt disease and is involved in age-related macular degeneration. The mutation changes a glycine to an alanine in exon 17, but in addition the change is at a splice acceptor site. A model of human splice acceptors built using information theory was used to identify acceptor sites in this region. The acceptor sites are shown by horizontal sets of letters called sequence walkers (Nucl. Acids Res., 25: 4408-4415, 1997). There is a strong 11.6 bit acceptor at the end of exon 17 of the wild type sequence (top) and a relatively weak cryptic acceptor of 3.9 bits 3 bases downstream. In the mutant sequence (bottom), the 11.6 bit acceptor has been degraded to 9.7 bits while the 3.9 bit cryptic acceptor has been increased to 9.8 bits. The two acceptors probably compete equally well for spliceosome binding, so that the mutation either causes a missense (G863A) or a deletion of the amino acid due to activation of the cryptic acceptor. See https://alum.mit.edu/www/toms/ and https://alum.mit.edu/www/toms/walker for further information.
2000 February 20: The prediction was correct! A paper was published in which the authors measured the amount of RNA and the two forms were observed: Maugeri et al. Am J Hum Genet 1999 Apr;64(4):1024-1035. The citation and abstract are available at pubmed and at AJHG.
![]()

Schneider Lab
origin: 1998 May 21
updated: 2008 Nov 20
![]()