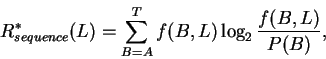 |
(10) |
This paper considers the relationship between
Rsequence and
Rfrequency.
For restriction enzymes cutting
genomes with equal numbers of the four bases randomly distributed,
Rsequence and
Rfrequency are equal.
For example, one commonly assumes that
HaeIII (GGCC; Roberts, 1983;
Rsequence = 8 bits)
cuts once in 256 bases (
Rfrequency = 8 bits).
This is not true for "skewed" genomes,
in which the frequencies of each base are significantly unequal.
For example, in a genome like that of bacteriophage T4
which is two-thirds A-T,
Rsequence for any tetramer is 7.7 bits.
Yet GGCC should occur once in every 1296 bases
(
( 1 / 6 )4;
Rfrequency = 10.3 bits)
and conversely AATT should occur once in every 81 bases
(
( 1 / 3 )4;
Rfrequency = 6.3 bits).
An alternative formula,