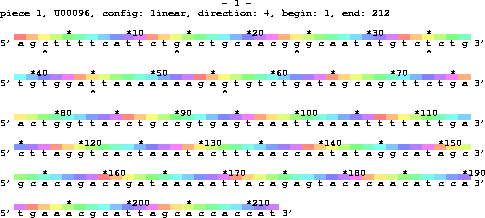
The live program produces a "LIster waVE" that cycles along the DNA sequence, usually to indicate the twist of B-form DNA (Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences, Nucl. Acids Res. 25: 4408-4415, 1997).
For example, a lister map of the first 212 E. coli bases
(U00096)
is:

The colors repeat precisely every 10.6 bp, according to the live parameter file, livep. This does not match the asterisk ("*") marks placed by Lister every 10 (and 5) bases, so the colors shift by a little relative to the asterisk. Compare the colors at 3, 14, 24, 35, 45 and 56 (marked with "^"). Each time the position of "yellowish" is a little later, because 10.6 is slightly greater than 10.0. The colors change slightly because 0.6+0.6 = 1.2, etc.
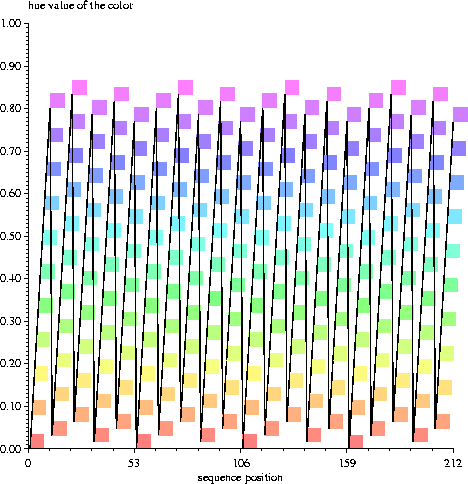
Above is a graph created from the live color marks file, livemarks. The hues are computed from the distance from the first position divided by 10.6. Each square corresponds to one of the bases in the sequence of the first figure, but the vertical position indicates the hue.
Postscript color hues run from 0 to 1, but 0 and 1 are both red and one doesn't get a good spectrum. So I alter the range to run between 0.0 and 0.85, which gives a nice spectrum from red to violet. The colors are only 50% saturated so that the color wave does not overly draw attention to itself.
Since 10.6*5 = 53.0 exactly, but not for smaller multipliers, the colors start to repeat at base 53. As can be seen in the plot, the colors shift slightly every 10 bases.
Publication that uses this technique: Discovery of Fur binding site clusters in Escherichia coli by information theory models NAR 35: 6762 (2007), Figure 5.
![]()

Schneider Lab
origin: 1999 June 2
updated: 2012 Oct 26
![]()