Control of Genetic Systems in Prokaryotes and Eukaryotes
Genetic Control in Prokaryotes
Prokaryotes have two levels of metabolic control
- Vary the numbers of specific enzymes made (regulation of gene expression)
- Slow, but can have a dramatic effect on metabolic activity
- Regulate enzymatic pathways (feedback inhibition, allosteric control)
- Rapid and can be fine-tuned, but if the enzyme system does not have
this level of control, then it is useless
Prokaryotes are "simple," single celled organisms, so they have
"simple" systems
- Genes are grouped together based on similar functions into functional
units called operons
- MANY GENES UNDER ONE CONTROL!!!
- There is one single on/off switch for the genes
lac operon in E. coli
- Function - to produce enzymes which break down lactose
(milk sugar)
- lactose is not a common sugar, so there is not a great need for these
enzymes
- when lactose is present, they turn on and produce enzymes
- Two components - repressor genes and functional genes
- Three functional genes:
- lacZ produces B-galactosidase. This enzyme hydrolyzes the
bond between the two sugars, glucose and galactose
- lacY produces permease. This enzyme spans the cell
membrane and brings lactose into the cell from the outside environment.
The membrane is otherwise essentially impermeable to lactose.
- lacA produces B-galactosidase transacetylase. The
function of this enzyme is still not known.
- Promoter (P) - aids in RNA polymerase binding
- Operator (O) - "on/off" switch - binding site for the
repressor protein
- Repressor (lacI) gene
- Repressor gene (lacI) - produces repressor protein w/ two
binding sites, one for the operator and one for lactose
- The repressor protein is under allosteric control - when not bound
to lactose, the repressor protein can bind to the operator
- When lactose is present, an isomer of lactose, allolactose, will
also be present in small amounts. Allolactose binds to the
allosteric site and changes the conformation of the repressor
protein so that it is no longer capable of binding to the operator
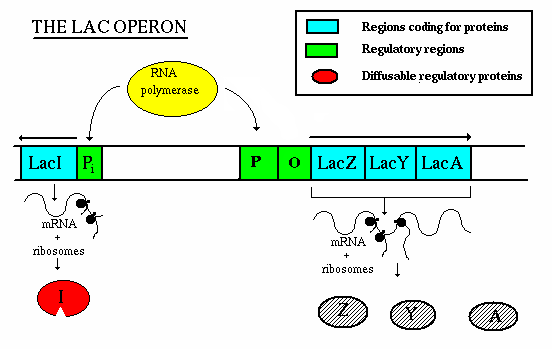
Operation - If lactose is not present:
- the repressor gene produces repressor, which binds to the operator. This
blocks the action of RNA polymerase, thereby preventing transcription.
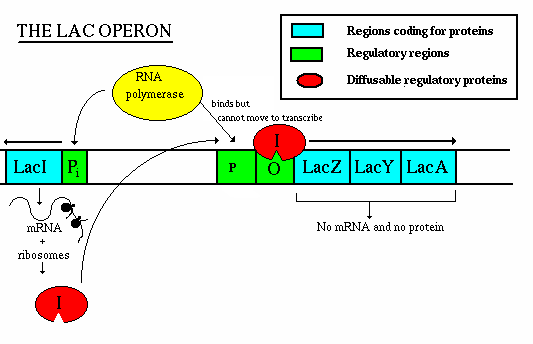
Operation - if lactose is present:
- the repressor gene produces repressor, which has a site for binding
with allolactose.
- The allolactose/repressor compound is incapable of binding w/ the
operator, so the RNA polymerase is uninhibited
- once the concentration of lactose decreases, the repressor-allolactose
complex falls apart and transcription is again inhibited
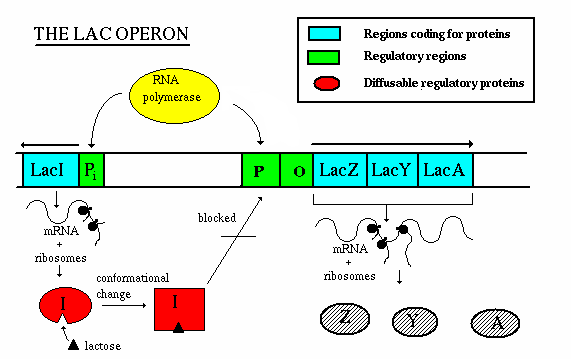
The lac operon is an example of an inducible operon - it is
normally off, but when a molecule called an inducer is present, the
operon turns on.
The trp operon is an example of a repressible operon - it is
normally on but when a molecule called a repressor is present the operon
turns off.
It Gets More Complicated - the lac Operon Revisited
It is not enough for lactose to be present to induce the lac operon
- Glucose is the sugar of choice of E. coli and if glucose is in
supply, then the bacteria will preferentially break down glucose over
lactose
- If glucose is present, the lac operon will be repressed - how does
this happen you ask?
- RNA polymerase has a low affinity for the promter of the lac operon unless
helped by a regulatory proten - cAMP receptor protein (CRP)
- CRP only becomes activated if the concentration of cyclic AMP (cAMP) is
high
- Glucose inhibits the formation of cAMP
- If the concentration of glucose is high, the concentration of cAMP is
low
- If the concentration of glucose is low, the concentration of cAMP is
high
- Therefore, if the concentrations of glucose and lactose are high, the
concentration of cAMP will be low, CRP will not be activated, RNA polymerase
will not be able to bind well to the promoter, and the operon will be
operating at a very low level (i.e. almost off)
- However, if the concentrations of glucose is low and lactose is high, the
concentration of cAMP will be high, CRP will be activated and bind to the
DNA which will promote RNA polymerase binding and initiate transcription

trp Operon - and example of a repressible operon
- five genes (trpA, trpB, trpC, trpD, and trpE) involved in
the production of the amino acid tryptophan
- another gene (trpR) produces an inactive repressor protein
- accumulation of the end product (tryptophan) represses synthesis of the
enzymes
- tryptophan binds to the inactive repressor protein at an allosteric
site
- the conformation changes and the repressor + tryptophan complex binds
to the operator, repressing the operon
- tryptophan can accumulate due to internal production or from external
sorces
- remember, E. coli is found in the intestines of humans so if
you eat a tryptophan-rich meal, this will accumulate in the bacteria and
turn off the operon
- why waste resources when a supply of this amino acid is readily
available?

Gene Control in Eukaryotes
Much more complex - take humans for example
- Every cell (except gametes) have the same DNA, with the same information
- This is known as genetic totipotency
- Almost all eukaryotic genes must be shut off in order to allow for
cell normal function (a liver cell cannot have genes for lung cells
running, not can it?)
- Usually, every gene has more than one gene regulator (all of which must be
on for the gene to function)
The latest
estimates are that a human cell, a eukaryotic cell, contains approximately
35,000 genes.
- Some of these are expressed in all cells all the time. These so-called
housekeeping genes are responsible for the routine metabolic functions (e.g.
respiration) common to all cells.
- Some are expressed as a cell enters a particular pathway of
differentiation.
- Some are expressed all the time in only those cells that have
differentiated in a particular way. For example, a plasma cell expresses
continuously the gene for the antibody it synthesizes.
- Some are expressed only as conditions around and in the cell change. For
example, the arrival of a hormone may turn on (or off) certain genes in that
cell.
How is gene expression regulated?
There are several methods used by eukaryotes.
- Transcription Control
- The most common type of genetic regulation
- Turning on and off of mRNA formation
- Post-Transcriptional Control
- Regulation of the processing of a pre-mRNA into a mature mRNA
- Translational Control
- Regulation of the rate of Initiation
- Post-Tranlational Control
- Regulation of the modification of an immature or inactive protein to
form an active protein

Transcriptional Control
 This is where a molecule of RNA polymerase II (pol II) binds. Pol
II is a complex of some 10 different proteins (shown in the figure in yellow
with small colored circles superimposed on it). The start site is where
transcription of the gene into mRNA begins.
The basal promoter contains a sequence of 7 bases (TATAAAA) called the TATA
box (this is very similar to the -10 box or Pribnow box found in
prokaryotes) . It can be bound by Transcription Factor IID (TFIID
read T F 2 D) which is a complex of some 10 different proteins including
This is where a molecule of RNA polymerase II (pol II) binds. Pol
II is a complex of some 10 different proteins (shown in the figure in yellow
with small colored circles superimposed on it). The start site is where
transcription of the gene into mRNA begins.
The basal promoter contains a sequence of 7 bases (TATAAAA) called the TATA
box (this is very similar to the -10 box or Pribnow box found in
prokaryotes) . It can be bound by Transcription Factor IID (TFIID
read T F 2 D) which is a complex of some 10 different proteins including
- TATA-binding
protein (TBP), which recognizes and binds to the TATA box
- other protein factors which bind to TBP - and each other - but not to the
DNA.
The basal or core promoter is found in all protein-encoding genes. This is in
sharp contrast to the upstream promoter whose structure and associated binding
factors differ from gene to gene (i.e. they are unique to each specific gene).
Although the figure is drawn as a straight line, the binding of transcription
factors to each other probably draws the DNA of the promoter into a loop.
Many different genes and many different types of cells share the same
transcription factors - not only those that bind at the basal promoter but even
some of those that bind upstream. What turns on a particular gene in a
particular cell is probably the unique combination of promoter sites and
the transcription factors that are chosen. To see how this all comes
together, click here.
The rows of lock boxes in a bank provide a useful analogy.
To open any particular box in the room requires two keys:
- your key, whose pattern of notches fits only the lock of the box assigned
to you (= the upstream promoter), but which cannot unlock the box without
- a key carried by a bank employee that can activate the unlocking mechanism
of any box (= the basal promoter) but cannot by itself open any box.
- Check out the movie "Matchstick Men" to see this in action
The complexes of hormones with their receptor represent one class of
transcription factor. Hormone "response elements", to which the
complex binds, are promoter sites. Link
to a discussion of these.
Just how do proteins bind to DNA?
DNA:Protein and Protein:Protein interactions are important for transcription
factor function. Note modular structure of transcription factors: one part of
the protein is responsible for DNA binding, another for dimer formation, another
for transcriptional activation (i.e. interaction with basal transcription
machinery).
Dimer formation adds an extra element of complexity and versatility. Mixing
and matching of proteins into different heterodimers and homodimers means that
three distinct complexes can be formed from two proteins.
Diverse in nature, but several common structures are found:
- Helix-turn-helix
(homeodomain) - three different planes of the helix are established and bind
to the grooves of the DNA
- Zinc
fingers - cystine and histidine residues bind to a Zn2+ ion,
looping the amion acid into a finger-like chain that will rest in the
grooves of DNA
- Leucine
zipper - dimers result from leucine residues at every other turn of the
a-helix. When the a-helical regions form a leucine zipper, the regions
beyond the zipper form a Y-shaped region that grips the DNA in a scissors-like
configuration

 Some
transcription factors ("Enhancer-binding protein") bind to regions of
DNA that are thousands of base pairs away from the gene they control. Binding
increases the rate of transcription of the gene.
Some
transcription factors ("Enhancer-binding protein") bind to regions of
DNA that are thousands of base pairs away from the gene they control. Binding
increases the rate of transcription of the gene.
Enhancers can be located upstream, downstream, or even within the gene they
control.
How does the binding of a protein to an enhancer regulate the transcription
of a gene thousands of base pairs away?
One possibility is that enhancer-binding proteins - in addition to their
DNA-binding site, have sites that bind to transcription factors ("TF")
assembled at the promoter of the gene.
This would draw the DNA into a loop (as shown in the figure).
- Enhancers can work even if thier normal 5' to 3' orientation is flipped
- Enhancers can work even if they are moved to a new location
- Regulatory sequences with similar characteristics, but the opposite
effect, exist. These are called silencers.
Silencers are control regions of DNA that, like enhancers, may be located
thousands of base pairs away from the gene they control. However, when
transcription factors bind to them, expression of the gene they control is
repressed.
A problem:
As you can see above, enhancers can turn on promoters of genes located
thousands of base pairs away. What is to prevent an enhancer from
inappropriately binding to and activating the promoter of some other gene in the
same region of the chromosome?
One answer: an insulator.
Insulators are
- stretches of DNA (as few as 42 base pairs may do the trick)
- located between the
- enhancer(s) and promoter or
- silencer(s) and promoter
of adjacent genes or clusters of adjacent genes.
Their function is to prevent a gene from being influenced by the activation
(or repression) of its neighbors.
 Example:
Example:
The enhancer for the promoter of the gene for the delta chain of the gamma/delta
T-cell receptor for antigen (TCR) is located close to the promoter for
the alpha chain of the alpha/beta TCR (on chromosome 14 in humans). A T
cell must choose between one or the other. There is an insulator between the
alpha gene promoter and the delta gene promoter that ensures that activation of
one does not spread over to the other.
All insulators discovered so far in vertebrates work only when bound by a
protein designated CTCF ("CCCTC binding factor"; named for a
nucleotide sequence found in all insulators). CTCF has 11 zinc fingers.
Another example: In mice (and humans), only the allele for insulin-like
growth factor 2 (Igf2) inherited from one's father is active; that
inherited from the mother is not - a phenomenon called imprinting.
The mechanism: the mother's allele has an insulator between the Igf2 promoter
and enhancer. So does the father's allele, but in his case, the insulator has
been methylated. CTCF can no longer bind to the insulator, and so the enhancer
is now free to turn on the father's Igf2 promoter.
Post-Transcriptional Control
All the primary transcripts produced in the nucleus must undergo processing
steps to produce functional RNA molecules for export to the cytosol. We shall
confine ourselves to a view of the steps as they occur in the processing of pre-mRNA
to mRNA.

The steps:
- Synthesis of the cap. This is a modified guanine (G) which is
attached to the 5' end of the pre-mRNA as it emerges from RNA polymerase II
(RNAP II). The cap protects the RNA from being degraded by enzymes that
degrade RNA from the 5' end.
- Step-by-step removal of introns present in the pre-mRNA and
splicing of the remaining exons. This step is required because most
eukaryotic genes are split. It takes place as the pre-mRNA continues to
emerge from RNAP II.
- Synthesis of the poly(A) tail. This is a stretch of adenine (A)
nucleotides. When transcription is complete, the transcript is cut at a site
(which may be hundreds of nucleotides before its end), and the poly(A) tail
is attached to the exposed 3' end. This completes the mRNA molecule, which
is now ready for export to the cytosol. (The remainder of the original
transcript is degraded and the RNA polymerase leaves the DNA.)
Most eukaryotic genes are split into segments. In decoding the open reading
frame of a gene for a known protein, one usually encounters periodic stretches
of DNA calling for amino acids that do not occur in the actual protein product
of that gene. Such stretches of DNA, which get transcribed into RNA but not
translated into protein, are called introns. Those stretches of DNA that
do code for amino acids in the protein are called exons. Examples:
- The gene for one type of collagen found in chickens is split into 52
separate exons.
- The gene for dystrophin, which is mutated in boys with muscular
dystrophy, has 79 exons.
- even the genes for rRNA and tRNA are split.
In general, introns tend to be much longer than exons. An average eukaryotic
exon is only 140 nts long, but one human intron stretches for 480,000
nucleotides!
The cutting and splicing of mRNA must be done with great precision. If even
one nucleotide is left over from an intron or one is removed from an exon, the reading
frame from that point on will be shifted, producing new codons specifying a
totally different sequence of amino acids from that point to the end of the
molecule (which often ends prematurely anyway when the shifted reading frame
generates a STOP codon).
The removal of introns and splicing of exons is done with the spliceosome.
This is a complex of several snRNA molecules and some 145 different
proteins.
The introns in most pre-mRNAs begin with a GU and end with an AG. Presumably
these short sequences assist in guiding the spliceosome.
Translational Control
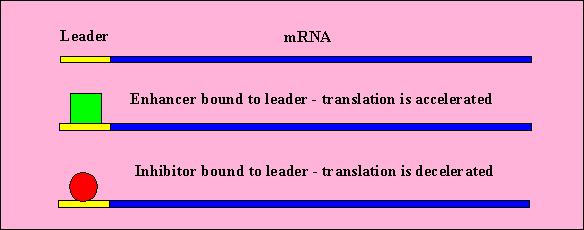
- There are regions on the beginning of mRNA which do not code for proteins.
These are the leaders.
- Proteins and other molecules can bind to the leader which can enhance
or restrict ribosome binding (and thus translation)
- mRNA molecules in cytoplasm may be degraded and recycled to make more RNA.
- This varies the amount of gene product that is produced (as a mRNA that's
degraded quickly won't express much protein).
Post-Translational Control










 Some
transcription factors ("Enhancer-binding protein") bind to regions of
DNA that are thousands of base pairs away from the gene they control. Binding
increases the rate of transcription of the gene.
Some
transcription factors ("Enhancer-binding protein") bind to regions of
DNA that are thousands of base pairs away from the gene they control. Binding
increases the rate of transcription of the gene.
 Example:
Example:


