
|
The
Consensus Sequence
by
Tom Schneider |
|

|
The
Consensus Sequence
by
Tom Schneider |
|
The concept of a consensus sequence is pervasive in molecular biology. It is a primitive concept that can and should be replaced by two concepts:
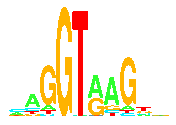

A poster describes how sequence logos and walkers form a complete replacement for consensus sequences.
This paper discusses the issues in detail:
|
T. D. Schneider,
Consensus Sequence Zen,
Applied Bioinformatics, 1 (3), 111-119, 2002. |
| See the oxyr, baseflip and repan3 papers for the science you WILL MISS if you use consensus sequences! |
Given that these concepts are well documented, already a good number of years ago, and given that consensus sequences are dangerous to use, scientific publications dated on or after 2001 May 1 that use the concept of a consensus sequence, when a sequence logo or a sequence walker would have been appropriate, will be placed on this page.
R. Swem, Sylvie Elsen, Terry H. Bird, Danielle L. Swem,
Hans-Georg Koch, Hannu Myllykallio, Fevzi Daldal, Carl E. Bauer,
The RegB/RegA Two-component Regulatory System Controls Synthesis of
Photosynthesis and Respiratory Electron Transfer Components in Rhodobacter
capsulatus.
Journal of Molecular Biology,
309(1): 121-138 (doi:10.1006/jmbi.2001.4652), May 2001.
"This high degree of sequence conservation raises the issue of whether or not
there is a consensus DNA-binding site that is recognized by RegA homologs ..."
This case demonstrates confusing the model (a consensus sequence)
with "reality" (a DNA-binding site).
 as of 2001 May 21.
as of 2001 May 21.
Patel PH, Suzuki M, Adman E, Shinkai A, Loeb LA.
Prokaryotic DNA polymerase I: evolution, structure, and "base flipping"
mechanism for nucleotide selection.
J Mol Biol. 2001 May 18;308(5):823-37.
Figure 3 gives
MOTIF B as
RpxxKxxxhGhhY
but the text gives
RRxhKhhNFGhhY
!
Likewise,
Figure 3 gives
MOTIF C as
hhxhHDxhxx
but the text gives says it contains
HDE!
This is a clear demonstration of how one can get into trouble.
 as of 2001 May 22.
as of 2001 May 22.
The crystal structure of a heptameric archaeal Sm protein:
Implications for the eukaryotic snRNP core.
Mura C, Cascio D, Sawaya MR, Eisenberg DS.
Proc Natl Acad Sci U S A. 2001 May 8;98(10):5532-7.
 as of 2001 May 23.
as of 2001 May 23.
Proc Natl Acad Sci U S A 2001 Aug
28;98(18):10085-9
Human DNA replication initiation factors, ORC and MCM,
associate with oriP of Epstein-Barr virus.
Chaudhuri B, Xu H, Todorov I, Dutta A, Yates JL.
Pubmed.
"ORC was first discovered in budding yeast as a six-member complex that binds in an ATP-dependent
manner to a consensus sequence present at yeast replication origins (30)."
Of course ORC binds to binding sites that are modeled by a consensus.
It does not bind to the model!
 as of 2001 Oct 2
as of 2001 Oct 2
Science 2001 Nov
2;294(5544):1098-102
Structural basis for recognition of the intron branch
site RNA by splicing factor 1.
Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango
S, Sprangers R, Zanier K, Kramer A, Sattler M.
Pubmed.
 as of 2001 Nov 8
as of 2001 Nov 8
Categorization and characterization of transcript-confirmed constitutively and
alternatively spliced introns and exons from human.
Hum Mol Genet. 2002 Feb 15;11(4):451-64.
Clark F, Thanaraj TA.
Pubmed PMID: 11854178.
The authors used sequence logos
 but used the term 'consensus sequence' all over the place
and did not use the individual information or walker methods.
but used the term 'consensus sequence' all over the place
and did not use the individual information or walker methods.
 as of 2002 March 28
as of 2002 March 28
Biochem Biophys Res Commun 2002 Mar
8;291(4):979-86
Evidence for "pre-recruitment" as a new mechanism of
transcription activation in Escherichia coli: the large excess
of SoxS binding sites per cell relative to the number of
SoxS molecules per cell.
Griffith KL, Shah IM, Myers TE, O'Neill MC, Wolf RE Jr.
Pubmed PMID: 11866462.
 as of 2002 April 23.
as of 2002 April 23.
J Virol 2001 Jun;75(12):5567-75
Retroviral constitutive transport element evolved
from cellular TAP(NXF1)-binding sequences.
Zolotukhin AS, Michalowski D, Smulevitch S, Felber BK.
Pubmed PMID: 11356964.
 as of 2002 May 16
as of 2002 May 16
Science 2002 May 10;296(5570):1132-6
A complex with chromatin modifiers that occupies E2F- and Myc-responsive
genes in G0 cells.
Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y.
Pubmed PMID: 12004135 .
 as of 2002 May 16
as of 2002 May 16
Rebeiz M, Reeves NL, Posakony JW.
SCORE: A computational approach to the identification of cis-regulatory
modules and target genes in whole-genome sequence data.
Proc Natl Acad Sci U S A. 2002 Jul 23;99(15):9888-93.
PMID: 12107285.
 as of 2002 July 31
as of 2002 July 31
James LC, Roversi P, Tawfik DS.
Antibody multispecificity mediated by
conformational diversity.
Science 2003 Feb 28;299(5611):1362-7
PMID: 12610298.
See figure 4.
 as of 2003 March 12
as of 2003 March 12
Lavrrar JL, McIntosh MA.
Architecture of a Fur binding site: a comparative analysis.
J Bacteriol. 2003 Apr;185(7):2194-202.
PMID: 12644489.
The abstract starts off:
"Fur ... recognizes a 19-bp consensus site ..."
This is
Confusing a model with reality.
 as of 2003 May 25
as of 2003 May 25
Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J,
Waterston R, Cohen BA, Johnston M.
Science. 2003 Jul 4;301(5629):71-6.
Finding functional features in Saccharomyces genomes by phylogenetic
footprinting.
PMID: 12644489.
Lots of "sequence motifs" - a terrible model!!
 as of 2003 May 25
as of 2003 May 25
ERROR MADE WITH CONSENSUS SEQUENCE
Prog Nucleic Acid Res Mol Biol. 1990; 38: 137-64.
Structure-function relationships in Escherichia coli promoter DNA.
Horwitz MS, Loeb LA.
PMID: 2183292.
 as of 2004 Jan 22
as of 2004 Jan 22
On page 141 of this paper the authors state:
For promoters of sigma70 holoenzyme, a general rule is that mutations that decrese a promoters's agreement with the consensus decrease the frequency of transcription initiation, and mutations that increase consensus agreement increase initiation frequency (20). A prediction of this rule is that the consensus sequence represents the most effecient promoter.So far so good. This should be true according to individual information theory. They continue:
The correctness of this rule is demonstrated by the success of an algorithm correlating consensus agreement and promoter strength in vitro (46).This should be improved upon by using matrix methods. They continue:
Exceptions to this rule are infrequent, but may be of significance. One example is the promoter for the arabinose operon araBAD, in which an A-to-G change at -33 decreases consensus homology while increasing promoter strength (47).This is a controvertial proposal. Is it correct? There are two flaws in the above quoted paragraph:
| wild type | TGACGC |
| mutant | TGGCGC |
| consensus | TTGAGA |
Murakami KS, Masuda S, Darst SA.
Structural basis of transcription initiation: RNA polymerase
holoenzyme at 4 A resolution.
Science. 2002 May 17;296(5571):1280-4.
PMID: 12016306.
 as of 2004 Jun 8
The bacterial RNAP holoenzyme forms an initial closed promoter complex
by recognizing two hexamers of consensus DNA sequence ..."
Nope, they recognize a lot more sequences than just the consensus!
as of 2004 Jun 8
The bacterial RNAP holoenzyme forms an initial closed promoter complex
by recognizing two hexamers of consensus DNA sequence ..."
Nope, they recognize a lot more sequences than just the consensus!
Grifantini R, Sebastian S, Frigimelica E, Draghi M, Bartolini E, Muzzi
A, Rappuoli R, Grandi G, Genco CA.
Identification of iron-activated and -repressed Fur-dependent genes by
transcriptome analysis of Neisseria meningitidis group B.
Proc Natl Acad Sci U S A. 2003 Aug 5;100(16):9542-7.
PMID: 12883001 .
 as of 2004 Jun 29.
Fur forms a dimer together with ferrous iron and binds to a consensus
sequence (Fur-box) that overlaps the promoters of iron
regulated genes, resulting in the inhibition of transcription.
This is a good example of confusing a model with the reality.
The guilty party in this case is the
FINDPATTERN program, part of the
Wisconsin package
(Accelrys, San Diego).
as of 2004 Jun 29.
Fur forms a dimer together with ferrous iron and binds to a consensus
sequence (Fur-box) that overlaps the promoters of iron
regulated genes, resulting in the inhibition of transcription.
This is a good example of confusing a model with the reality.
The guilty party in this case is the
FINDPATTERN program, part of the
Wisconsin package
(Accelrys, San Diego).
Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K,
Koeppen H, Dixit VM.
The ubiquitin ligase COP1 is a critical negative regulator of p53.
Nature. 2004 May 6;429(6987):86-92.
PMID: 15103385 .
In the legend to Figure 5:
 as of 2004 Jul 9.
"Identification of a p53 consensus site on the COP1 promoter."
This is another example of confusing a model (consensus sequence)
with the reality (binding site).
as of 2004 Jul 9.
"Identification of a p53 consensus site on the COP1 promoter."
This is another example of confusing a model (consensus sequence)
with the reality (binding site).
Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M.
Regulation of gene expression by a metabolic enzyme.
Science. 2004 Oct 15;306(5695):482-4.
PMID: 15486299 .
In the legend to Figure 3:
 as of 2004 Oct 25.
"Consensus motif derived from targets confirmed by PCR."
No, the image is an (uncited!)
sequence logo
which is NOT a consensus sequence.
as of 2004 Oct 25.
"Consensus motif derived from targets confirmed by PCR."
No, the image is an (uncited!)
sequence logo
which is NOT a consensus sequence.
Wei G, Li AG, Liu X. J Biol Chem. 2005 Jan 20. Insights into selective activation of p53 DNA binding by c-Abl. PMID: 15661746 . "As a transcription factor, p53 recognizes a specific consensus DNA sequence ..." This is the protein recognizing a model, and, of course, most p53 sites are not the consensus.
Nucleic Acids Res. 2005 May 11;33(8):2726-33. 2005.
Common patterns in type II restriction enzyme binding sites.
Nikolajewa S, Beyer A, Friedel M, Hollunder J, Wilhelm T.
PMID: 15888729 .
Table 3 is full of consensus sequences.
The natural variation that they suppressed
ruins the point they want to make.
 as of 2006 Jan 12.
as of 2006 Jan 12.
If you would like to contribute a reference to this page, please follow these rules:
![]()

Schneider Lab
origin: 2001 Apr 23
updated: 2012 Apr 10
![]()