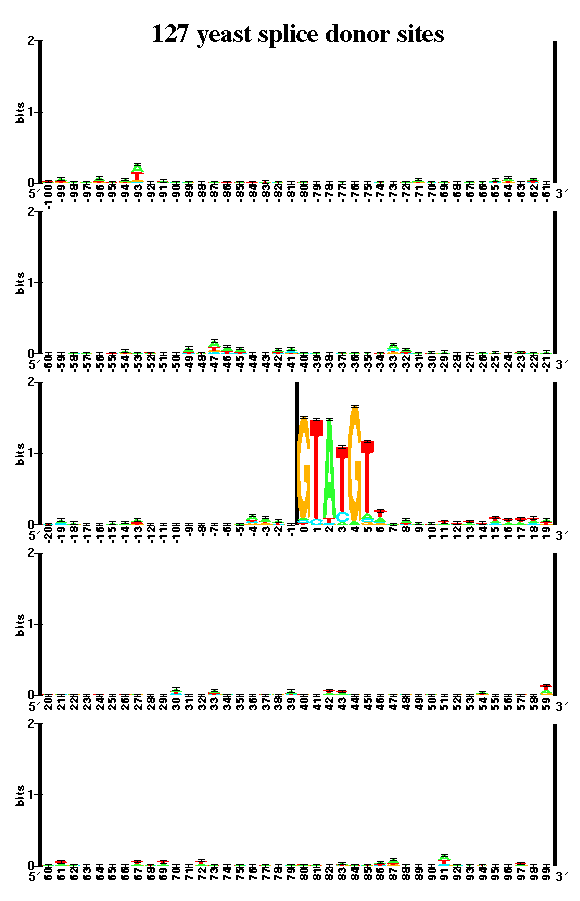
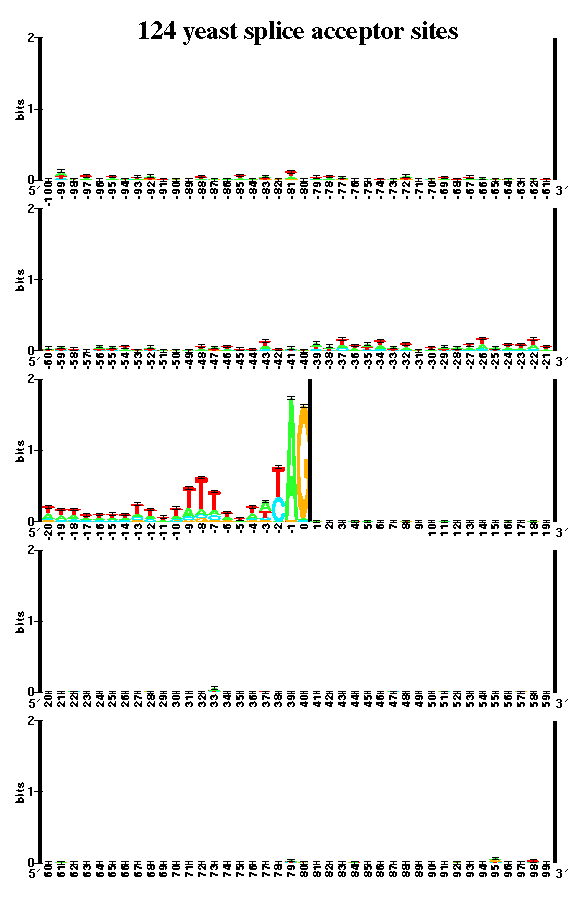
A sequence logo is made up of stacks of letters. Each stack contains the 4 bases. The height of each letter is proportional to its frequency at that position in the binding site, and the letters are sorted so that the most frequent one is on top. The height of the entire stack of letters is measured in bits of information, and represents the sequence conservation.
The database used for this analysis consists of the 124 introns reported in the complete yeast genome.
A published sequence logo for yeast appears in PNAS 95: 219-223, 1998, Evolution Relationship between "proto-splice sites" and intron phases: Evidence from dicodon analysis, Manyuan Long, Sandro J. de Souza, Carl Rosenberg, and Walter Gilbert



Schneider Lab.
origin: 1998 March 2
updated: 1998 March 2
