
abstract:
A graphical method is presented for displaying how binding proteins and other macromolecules interact with individual bases of nucleotide sequences. Characters representing the sequence are either oriented normally and placed above a line indicating favorable contact, or upside-down and placed below the line indicating unfavorable contact. The positive or negative height of each letter shows the contribution of that base to the average sequence conservation of the binding site, as represented by a sequence logo. These sequence `walkers' can be stepped along raw sequence data to visually search for binding sites. Many walkers, for the same or different proteins, can be simultaneously placed next to a sequence to create a quantitative map of a complex genetic region. One can alter the sequence to quantitatively engineer binding sites. Database anomalies can be visualized by placing a walker at the recorded positions of a binding molecule and by comparing this to locations found by scanning the nearby sequences. The sequence can also be altered to predict whether a change is a polymorphism or a mutation for the recognizer being modeled.
Tue, Sep 30, 1997
12:00 noon.
NOTE TIME CHANGE
(previous time was 11 am)
8th Floor Conference Room of Building 38A
National Library of Medicine
Bethesda, MD.
See the new parking scheme on the NIH campus.
We plan to have lunch afterwards.
Note: This is the same talk as https://alum.mit.edu/www/toms/bitcs/schneider1997oct1.html and https://alum.mit.edu/www/toms/bitcs/schneider1997oct14.html
Seminar arrangements:
David Landsman
(landsman@quagga.nlm.nih.gov)
(301) 496-2477 x223
This announcement is at https://alum.mit.edu/www/toms/bitcs/schneider1997sep30.html
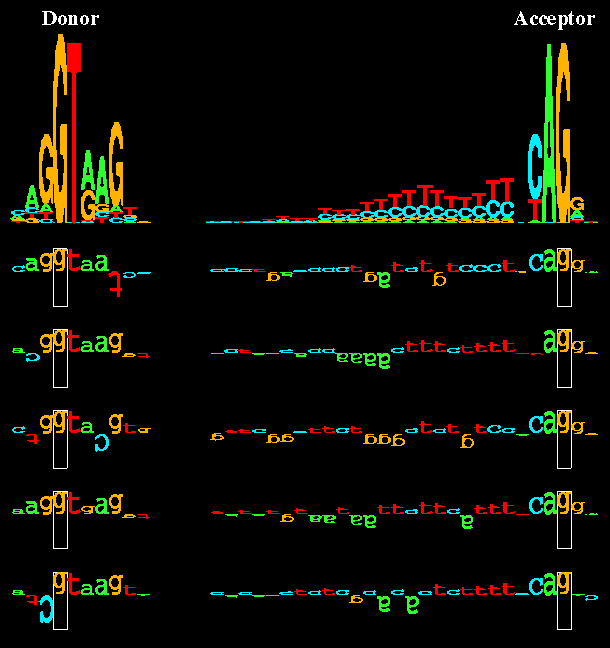
Sequence logos and walkers. At the top of the figure sequence logos for
human donor and acceptor sites depict the DNA sequence conservation by stacks
of letters. Within each stack the height of each letter is proportional to
the base frequency at that position in the binding site. Below the sequence
logos, five examples of individual binding sites are shown as walkers. The
height of each letter shows its contribution to the average sequence
conservation in the sequence logo on the top.
(This figure will be the cover of Nucleic Acids Research volume 25,
issue 21.)


Schneider Lab.
origin: 1997 September 15
updated: 1999 November 24
